Bacteria: How do they resist?
What is it?
When is the last time you were prescribed antibiotics? Aren't they simply amazing? Painful, unpleasant, potentially life-threatening infections cleared up in a matter of days: pneumonia, cellulitis, abscesses, urinary tract infections, bacterial meningitis, etc. Extraordinary!
But what about when the drugs don't work? Antibiotic resistance is one of the most significant threats to global public health that we face today, and fear for the future.
Bacteria have evolved a myriad of ways to fight back against our attempts to kill them with antibiotics. This includes production of β-lactamase, a class of enzymes that are able to degrade antibiotics containing a β-lactam ring. β-lactam antibiotics include penicillin, carbapenems and monobactams.
Where did the structure come from?
 This is the structure of a β-lactamase from Staphylococcus aureus. S. aureus is responsible for a variety of skin infections and is a major contributor to post-surgical infections in hospitals.
This is the structure of a β-lactamase from Staphylococcus aureus. S. aureus is responsible for a variety of skin infections and is a major contributor to post-surgical infections in hospitals.
What does it look like?
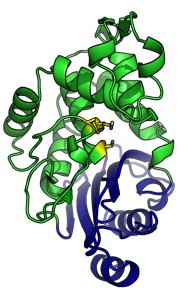
S. aureus β-lactamase is a small protein made up of two closely associated domains (coloured green and blue.) The enzyme active site is located in the interface between the two domains, and contains three catalytically important residues (serine, lysine and glutamate, highlighted in yellow) that are responsible for hydrolysis of the β-lactam ring.
This structure is Protein Data Bank ID 3BLM
References
- Herzberg, O. & Moult, J. (1987). Bacterial resistance to beta-lactam antibiotics: crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.5 Å resolution. Science. 236:694-701.
- Herzberg O. (1991). Refined crystal structure of β-lactamase from Staphylococcus aureus PC1 at 2.0 Å resolution. J Mol Biol. 217:701-719.