Munc18: No Munc-eying around with our molecular transport machinery!
What is it?
The transport of chemicals or proteins across the membranes of cells is a vital part of life. Chemicals may need to be released in order to send a signal to the cell next door, or, sometimes, a protein must be relocated from inside the cell to sit on the cell membrane so it can do its job.
For this reason, the Sec/Munc18 (SM) family of proteins — involved in fusion between two membranes — are an essential for this molecular transport and play a key part of healthy cell function. These SM proteins are involved in neurotransmitter release in the brain and in the maintenance of glucose levels (i.e. by relocating the glucose transporter to the cell surface so it can pump glucose into the cell).
Within our cells, chemicals or molecules can be isolated from the rest of the cell contents by containing them within small membrane encapsulated vesicles. But the molecular mechanisms behind how proteins, including SM, coordinate the fusion of these vesicle membrane to the outer membrane of the cell, so that neurotransmitters or the glucose transporter can do their thing, is not understood.
Crystal structures of SM proteins alone, and in complex with their partner proteins, are helping us to decode the complex mechanisms of the membrane fusion event. And the importance of research in this area is essential for the development of new treatments for neurological diseases or diabetes. Indeed, Randy W. Schekman, Thomas C. Südhof, and James E. Rothman were awarded the 2013 Nobel Prize in Physiology or Medicine for their discoveries on the protein machinery that regulates vesicle traffic, including the SM proteins! You can read more about their Nobel Prize and their research here.
What does it look like?
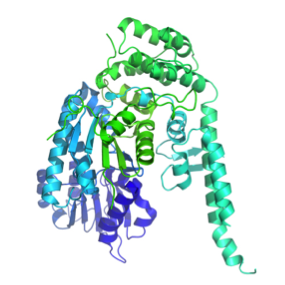
SM proteins, such as Munc18-1 (or Munc18a) shown here, consist of three separate domains that fold into a U-shaped or arch-shaped architecture. Image generated by Pymol (using the coordinates from the Protein Data Bank (accession code: 3PUJ).
Where did the structure come from?
This is the structure of rat Munc18-1 that was solved in complex with a portion of its partner protein, syntaxin (Sx). The structure was published in 2011 PNAS by a team of researchers from the Institute for Molecular Bioscience, UQ, Australia.
A total of 12 SM structures have been determined, either alone or complexed to a truncated protein partner or short peptide sequence. Click here for a review on the current structural information of SM proteins.