Superbug superpowers
What is it?
For this, the second installment in mechanisms of bacterial antibiotic resistance, (read the first here) we turn to Penicillin Binding Protein 2a (PBP2a) from methicillin resistant Staphylococcus aureus or MRSA.
Fuelled by sensationalist headlines such as "Superbug crisis worse than feared" and "MRSA out of control and getting stronger", MRSA is now synonymous with the most feared of resistant bacterial infections. But what do we know about how it can side-step antibiotic action?
As the name indicates, MRSA is resistant to the β-lactam antibiotic methicillin. Methicillin – like other β-lactams – mimics a substrate of a penicillin binding protein (PBP). Bacterial PBPs are necessary for cell wall assembly, and antibiotics such as methicillin disrupt cell wall formation by inhibiting normal substrate binding to PBPs.
MRSA can resist methicillin because they have acquired a mecA gene that produces a PBP variant – PBP2a – that simply doesn't bind β-lactam antibiotics very well. As a result, the PBPs evade methicillin inhibition and continue to build cell walls normally.
Where did the structure come from?
This is the structure of a mecA gene encoded PBP2a from MRSA strain 27r [1].
What does it look like?
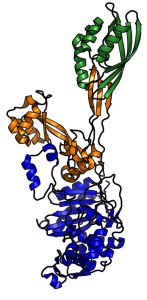 PBP2a is a large multi-domain protein; the transpeptidase domain (blue) and penicillin binding domain (yellow) work together to assist proper cell wall synthesis. Comparison of methicillin-sensitive and methicillin-resistant PBPs revealed that methicillin-resistant PBP2a has evolved a distorted active site in its penicillin-binding domain that disrupts interaction with β-lactams, protecting the protein, and thus the bacterium, from antibiotic inhibition.
PBP2a is a large multi-domain protein; the transpeptidase domain (blue) and penicillin binding domain (yellow) work together to assist proper cell wall synthesis. Comparison of methicillin-sensitive and methicillin-resistant PBPs revealed that methicillin-resistant PBP2a has evolved a distorted active site in its penicillin-binding domain that disrupts interaction with β-lactams, protecting the protein, and thus the bacterium, from antibiotic inhibition.
This structure is Protein Data Bank ID 1VQQ
Reference:
[1] Lim and Strynadka. Nature Structural Biology 9: 870-876, 2002.